Molecular Variability of Citrus tristeza virus Determined by
advertisement

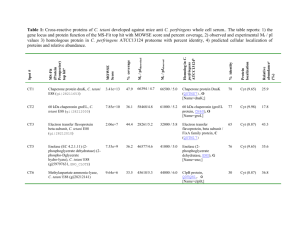
Molecular Variability of Citrus tristeza virus Determined by Assymmetric PCR-ELISA and SSCP Silvija Černi1, Vera Martins2, Mladen Krajačić1, Dijana Škorić1 ,Gustavo Nolasco2 of Zagreb, Faculty of Science, Department of Biology, Marulićev trg 9A, Zagreb, 2Universidade do Algarve, Faculdade de Engenharia de Recursos Naturais, Gampus de Gambelas, Faro, Portugal 1University Dipl. ing. Silvija Černi, University of Zagreb, Faculty of Science, Department of Biology, Marulićev trg 9A, 10000 Zagreb, Croatia Tel. +(385)14843851 Fax. +(385)14844001 E-mail: scerni@botanic.hr 1 Citrus tristeza Closterovirus (CTV) is one of the most important viral pathogens of citrus. CTV virions are phloem-limited flexuous filaments of 2000x11 nm in size. With its monopartite, single-stranded, positive-sense RNA genome of 19.3 kb organized into 12 open reading frames (ORFs), CTV is considered to be the largest single-stranded RNA plant virus characterized so far. Aphids naturally spread the virus but it is also easily transmitted by grafting. Depending on a virus strain, scion cultivar and rootstock, CTV can induce one of the three main syndromes: “quick decline”, “stem pitting” and “seedling yellows”, but some mild strains can go unnoticed because of the lack of visible symptoms. In nature, CTV usually exists as a mixture of strains. Due to a low concentration and seasonal variations of the virus in field trees, the detection is difficult and it is necessary to use methods for sensitive and reliable CTV diagnosis and strain typing. Five CTV positive Croatian citrus samples have been strain-typed using asymmetric PCR ELISA and single strand conformation polymorphism (SSCP) analysis. CTV coat protein (CP) gene was amplified in an immunocapture asymmetric RT-PCR (IC/RT-PCR) and amplicons were hybridized with the eight CTV group-specific probes. In addition, SSCP analysis of CTV CP gene was performed. According to the probe-typing results, all five analyzed samples were mix-infected and showed homology to CTV phylogenetic groups 2 and M which represent mild, asymptomatic, CTV strains. One sample also showed homology to probe from group 4 and another to probe from group 3a. Groups 3a and 4 encompass very severe, “quick decline” strains and group 3a can additionally induce “stem pitting” syndrome. Multiple band patterns in SSCP profiles confirmed mixed-infectious in all analyzed samples. Probe-typing results were further corroborated by cloning and sequencing of IC/RT-PCR amplicons. Phylogenetic analysis was in accordance with the probe-typing groupclassification for all the samples. Neither of the amplicons, even from the same virus strain, had the same sequence. All the results obtained represent molecular confirmation of the considerable biological diversity of CTV found in Croatian citrus samples. 2 3