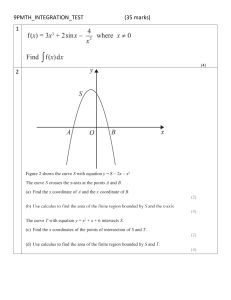
CHEM2211 End of Semester Practice Exam Questions 120 minutes + 15 minutes reading time Total of 120 marks (~ 1 mark per minute) 10 multiple choice questions followed by short/medium answer questions (in parts). Part 1. True/False and Multiple Choice Questions (covering all of Joe’s lectures) [10 Questions, 20 marks, 2 marks each] Q1.1 Which of the following is an example of a spontaneous process a) The conversion of graphite to diamond under standard conditions b) The movement of a molecule from an area of high chemical potential to an area of low chemical potential c) The conversion of ADP + Pi à ATP under standard conditions d) A chemical reaction which has a Keq = 0.8 Q1.2. Which of the following is an extensive property? a) b) c) d) e) f) Temperature Enthalpy Density Pressure Colour Chemical potential Q1.3. Which of the following techniques is most suitable for determining the melting temperature (Tm) of a protein? a) b) c) d) e) f) Protein X-ray crystallography Isothermal titration calorimetry AlphaFold2 Differential scanning calorimetry Small-angle x-ray scattering Surface plasmon resonance Q1.4. True or false? The universal binding isotherm (shown below) can be used to describe/model any binding interaction between a protein and a ligand. Q1.5. True or false? The concentration of Mg2+ needed to promote the folding of an RNA molecule is lower than the concentration of Na+ needed to promote RNA folding. Q1.6. Which of the following proteins does NOT use the energy released by the hydrolysis of ATP to drive a thermodynamically unfavourable process? a) GroEL b) Kinesin c) ATP Synthase (this uses movement of protons down electrochemical gradient to drive the production of ATP from ADP + Pi.) d) Heat shock protein 70 e) Maltose ABC transporter Q1.7 Here is some data comparing different binding interactions of a drug with two different proteins. Which of the following statements is TRUE?: a) The interaction between the drug and Protein 2 has a lower affinity than the interaction between drug and Protein 1. b) The interaction between the drug and Protein 2 has a higher KD than the interaction between the drug and Protein 1. c) Considering these two interactions, the specificity constant remains the same when the concentration of the drug is increased. d) The ratio of the KD between Protein 1 and the drug and KD of interaction of Protein 2 ! and the drug (i.e. !!,# ) changes as the concentration of the drug is increased. !,$ " e) The ratio of the fractional occupancy of Protein 1 and Protein 2 (i.e. "%&'()*+ # ) %&'()*+ $ changes as the concentration of the drug is increased. Q1.8 Which of the following statements is FALSE? a) Hydrophobic residues are typically important for conferring the affinity in proteinprotein interactions. b) Interface waters do not contribute to binding specificity in protein-protein interactions. c) Proteins that bind DNA tend to have positively-charged surfaces that interact with the negatively-charged DNA molecules. d) Interface waters can help to improve packing at protein-protein interfaces. e) SH2 domains typically bind peptides that contain a phosphotyrosine residue. Q1.9 True or False? If the KDs of two different binding interactions are the same, it means that the rate constants for association (kon) and rate of dissociation (koff) must be the same for these interactions. Q1.10 Which of the following is an example of allosteric regulation or an allosteric binding interaction? a) Competitive inhibition b) Binding of maltose to the maltose binding periplasmic binding protein. c) Binding of a phosphotyrosine-containing peptide to a SH2 domain d) Consecutive binding of O2 to the four heme molecules in Hemoglobin. e) A drug that binds two different protein targets (each with independent binding sites). Part 2. [Free Energy, Work & Equilibria] [20 Marks] 2.1 Fundamental Equations [9 MARKS] The following equations can be thought of as “fundamental equations” that describe the how the total internal energy (U) or enthalpy (H) change during most chemical reactions or processes: a) In the above equations, which term represents the heat exchange during the chemical reaction? [1 MARK] b) In the equations above, what does the term ∑𝑵 𝒊%𝟏 𝝁𝒊 𝒅𝒏𝒊 represent and what are μi and dni? What is the significance of the sum symbol (S) in this term? [4 MARKS] c) Describe a specific situation in which the above equations would not be sufficient for describing the changes in internal energy or enthalpy of a biological process [2 MARKS] d) With reference to the above equations, why do biologists and biochemists tend to talk in terms of changes in enthalpy (H) rather than changes in the internal energy of a system (U)? [2 MARKS] 2.2 Electron transport chain & ATP Synthase [6 MARKS] The production of ATP from ADP and Pi is thermodynamically unfavourable under standard conditions (i.e. ΔG > 0). With this in mind, describe how the proteins of the electron transport chain and ATP synthase enable the production of ATP in the cell. [6 MARKS] 2.3 Amino acid acid-base equilibria [5 MARKS] The carboxyl side chain of free L-Glutamate has a pKa is about 4. a) At pH 7, what is the ratio of deprotonated vs protonated glutamate (i.e. [Glu-]/[Glu])? (show your working) [2 MARKS] b) Some enzymes rely on having protonated Glu residues (i.e. glutamic acid) in their active site even at physiological pH (e.g. pH 7). Considering that Glu has a pKa of 4 in water, explain how it is possible to have a Glu residue in the active site of an enzyme that remains protonated at pH 7. [3 MARKS] Part 3 [Protein/RNA Folding] [40 marks] 3.1 [12 MARKS] Protein dynamics and protein folding can both be explained in terms of conformational energy landscapes. a) In reference to proteins, define the term conformational energy landscape. [4 MARKS] b) Explain how the conformational energy landscape of a protein can be used to describe the process of protein folding. [4 MARKS] c) Explain how the conformational energy landscape of a protein can be used to describe the dynamic nature of folded proteins. [4 MARKS] 3.2 Differential Scanning Calorimetry (Protein Stability) [13 MARKS] Joe has done a differential scanning calorimetry (DSC) experiment on his monomeric protein. Data is shown below: a) Briefly explain how a DSC instrument works and what information a DSC experiment can provide about protein unfolding. [4 MARKS] b) What is the melting temperature (Tm) of Joe’s protein? [1 MARK] c) Which part of the plot will tell Joe the ΔCp of the protein between unfolded and folded states? [1 MARK] d) Explain how can Joe calculate ΔH°unfolding and ΔS°unfolding at Tm from this data. [3 MARKS] Continued next page…. Joe runs a similar DSC experiment on a different protein and plots how ΔG°unfolding changes as a function of temperature: a) At what temperature is the protein most stable? [1 MARK] b) What is the Tm of this protein? [1 MARK] c) What is happening when the temperature is lowered below the temperature at which this protein is most stable? [2 MARKS] 3.3 Protein mis-folding and aggregation [7 MARKS] a) What is the difference between amorphous aggregation and amyloid fibrils.[2 MARKS] b) Name three situations when protein mis-folding and/or aggregation is commonly observed. Explain why protein misfolding or aggregation is common in these situations (your answer should explain the main drivers of protein aggregation). [5 MARKS] 3.4 Compare protein folding and RNA folding. What are similar about the two processes? How are they different? [8 MARKS] Part 4 [Molecular recognition] [40 marks] 4.1 [14 MARKS] Joe is looking at two interactions – one between a drug and on-target (Protein 1) and one between the same drug and an off-target (Protein 2). Data are shown below: a) What are the KDs of the two interactions? [2 MARKS] b) Calculate their corresponding ΔG°binding values (at 300 K) of the two interactions based on your answer to (a). Show your working. [4 MARKS] c) What is the minimum concentration of drug that should be used to ensure near-saturation (>95%) of on-target? [4 MARKS] d) Considering these two competing interactions, describe how the specificity of this interaction changes as [L] is increased from 10-11 M to 10-3 M. (you may want to show this using a plot) [4 MARKS] 4.2. Define and compare the following three models of protein-ligand binding: [10 MARKS] - Lock-and-key Induced-fit binding Conformational selection In your answer, for each model, include a plot showing how the energy landscape changes in the presence of the ligand. (Below is an example for the lock-and-key model). 4.3. With reference to the following data. Explain how isothermal titration calorimetry instrument works and what information can be obtained from an ITC experiment on an ideal, 1:1 binding interaction (and how it can be obtained). [10 MARKS] 4.5 Allostery [6 MARKS] Joe is studying a dimeric protein with two binding sites that bind the same ligand. The protein exists in two states: a low affinity state and a high affinity state. In the absence of ligand, both sites exist primarily in the low affinity state. Binding of the ligand to the first site switches the second site from the low affinity to the high affinity state. a) Is this an example of positive cooperativity or negative cooperativity? [1 MARK] b) In the context of protein-ligand interactions, describe what is meant by an ultrasensitive response. Describe how allosteric proteins can yield ultrasensitive responses and why this is important in a biological context. [5 MARKS] END OF PRACTICE EXAM