Deviation of amino acid utilization and correlation with G+C
advertisement

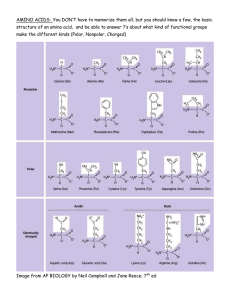
Deviation of amino acid utilization and correlation with G+C composition in bacterial genome Sajia Akhter, Hochul K Lee, Barbara Bailey, Peter Salamon, Robert Edwards Computational Science Research Center, San Diego State University 2008 Kullback-Leibler Divergence on Amino Acid Composition 0.09 Kullback-Leibler Divergence 0.08 The Kullback‐Leibler divergence (KLD) was calculated to compare the distribu4on of amino acids in different protein coding sequences as a measure of how much those sequences deviate from the standard. The Kullback‐Leibler divergence (KLD) was calculated for 372 whole bacterial genomes and for proteins in subsystems by As used here, Pi is the frequency of the ith amino acid in a given bacterial genome and Q i is the average frequency of ith amino acid calculated from all complete genomes. Non‐diverge Genome 0.07 0.06 Divergence of Amino Acid u4liza4on are not significantly different from the mean for all subsystems. 0.05 0.04 0.03 0.02 0.01 0 A mino A cids and Derivatives Carbohydrates Cell Divis ion and Cell Cycle DNA M etabolis m M embrane Trans port Nitrogen M etabolis m Phos phorus M etabolis m Protein M etabolis m RNA M etabolis m Sulfur M etabolis m Different Subsystems Possible Explanation Amino Acid Utilization for Divergence of Bifidobacterium adolescentis 0.16 0.14 Mean of KLD with SEM 0.12 Salmonella bongori 12149 Chlamydophila pneumoniae CWL029 Mean Frequency of Amino Acid Utilization Secondary metabolism has poor correla4on between GC content and amino acid composi4on – High level of horizontal gene transfer Limited (167) bacteria have this subsystem, and most of those have GC content between 40% and 60%. Predicting Amino Acid composition based on G+C content An explicit expression for the informa4on content is available once a surprisal/ devia4on analysis is carried out (Levine, 1978) m ln(Qi /Pi ) = "0 + # "r Ar (i) ! r=1 where Ar(i) are a set of m proper4es for the state i. For the devia4on of amino acid composi4on, since the interested property is only GC content, the model ! will be ln(Qi/Pi) = λ0 + λ (GC%) [eqn 1] Amino Acids and their GC Sensi4vity !"#$(-,$&%'(%)%*+(,$ 40,5%-(%(0$647$ !18,+9%(0$6:7$ ;5<0-(%(0$6;7$ 0.1 0.08 0.06 Amino Acid 0.04 0.02 a s ae ri te te og ae ac ot ch m ob er iro are significantly different from the mean for all subsystems. The differences are not restricted to one or few metabolic process but are across all subsystems. 0.3 0.25 0.2 $ Th Sp a ia ria te ac ob te ro ap m ia ri er ct ba eo te ot ro np lo si am Class of Bacteria Divergence of Amino Acid Utilization in different Subsystems The most Divergent Genome The most diverse Genome have low GC content (ranging from 22% to 28%) GC‐poor bacteria have few codons for alanine, glycine, proline, and arginine GC‐rich bacteria have few codons for phenylalanine, isoleucine, lysine, asparagine, and tyrosine Divergence of Amino Acid Utilization and G+C content G ia er er te ct ct ac ba ob eo te ot ro pr pr ap et el B D Ep i ha lp A ta es ut lic ba Fu so i ia ill cc ac rid ol M co st B no lo C e ia ia ia er ob ct or ba hl ei C no D C ya a ri ria te yd te ac ac am ob ob hl ng hi Sp C e ri ca et te ifi id ro qu te A av ac B Fl ac ob in ct A es a 0 Kullback-Leibler Divergence Nostoc sp. PCC 7120 Frequency of Amino acid Life S'le of Organism The organisms which have the most skewed amino acid composi4ons, are intracellular pathogens with a very limited ecological niche range and restricted lifestyle. Phylogene'c Effects There is a significant difference between amino acid u4liza4on in different phylogene4c groups of bacteria. Bacillus B-14905 The rela4onship between %GC and amino acid divergence is given by the equa4on y = 2(x‐0.5)2, where x is the %GC and y is the divergence of amino acid composi4on. Most subsystems has similar parabolic equa4on with high regression coefficient, which suggest that the DNA content and amino acid composi4on were related. 0.15 From eqn 1, Qi = Pi exp (λ0 + λ (GC%)) where, λ = fidng equa4on 1 with actual frequency λ0 = weighted average of G+C content Finally, Qi = Pi exp λ (GC% ‐ avg(GC%)) Previous Work According to Knight’s (2001) correla4on between Amino Acid and GC% Qi = λ0 + λ (GC%) Significance of Exponen'al rela'onship than Linear rela'onship Exponen4al rela4onship uses 1 parameter (λ) instead of 2 (λ and λ0) though the Regression coefficient (R^2) is almost same for both rela4onship. References 0.1 0.05 0 A mino A cids and Derivatives Carbohydrates Cell Divis ion and Cell Cycle DNA M etabolis m M embrane Trans port Nitrogen M etabolis m Phos phorus M etabolis m Protein M etabolis m RNA M etabolis m Different Subsystems Wigglesworthia glossinidia Borrelia garinii Mycoplasma mycoides Ureaplasma parvum serovar Buchnera aphidicola Mean Sulfur M etabolis m !"#$%&$&%'(%)%*+(,$ ./0$&1-20$ 3/0$&1-20$ =1+(%(0$6=7$ !18,+9%*$+*%?$6G7$ ">&,0%(0$6"7$ C50(>1+1+(%(0$6H7$ =&2+<,%*$+*%?$6@7$ I&-108*%(0$6I7$ !1>*%(0$6!7$ B>&%(0$6J7$ A%&,%?%(0$6A7$ =&2+<+'%(0$6K7$ B08*%(0$6B7$ L0<%(0$6L7$ C<-1%(0$6C7$ ;><-&%(0$6M7$ =<'%(%(0$6D7$ E+1%(0$6E7$ ;<>2,-25+($6F7$ Levine (1978), “Informa4on Theory Approach to Molecular Reac4on Dynamics” Annual Review of Physical Chemistry, 29(1):59 Knight (2001), “A simple model based on muta4on and selec4on explains trends in codon and amino‐acid usage and GC composi4on within and across genomes” Department of Ecology and Evolu4onary Biology, Princeton University, Princeton, NJ 08544, USA