Some of you may want to use Maple to make... a package named Networks that has many of the standard network...
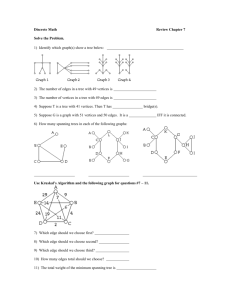
advertisement

Some of you may want to use Maple to make graph computations. Maple has
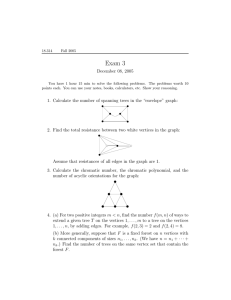
a
package named Networks that has many of the standard network algorithms.
From an x-window at one of the MSCC computers enter the command maple and
a
maple window will come up. Enter the command with(networks) to load the
networks package. You can find out what is in the package with the
command
?networks. To find out about any of the functions use the command
?networks(function).
Here is the output of the command ?networks:
HELP FOR: The Networks Package
SYNOPSIS:
- To use a networks function, either define that function alone using the
command with(networks, <function>), or define all networks functions using
the
command with(networks). Alternatively, invoke the function using the
long
form networks[<function>]. This long form notation is necessary
whenever
there is a conflict between a package function name and another
function used
in the same session.
- A network is represented by a graph consisting of vertices and edges.
The
edges may be directed, and loops and multiple edges are allowed. The
basic
routines in this package perform the manipulation of the underlying
graphs.
Various properties of the networks as outlined below can also be
computed.
In particular extensive computations for all terminal network
reliability
have been carried out using the routines present here.
- A graph is represented by a special Maple procedure of type GRAPH. A
graph
is created by the command new() or by commands such as
complete(), cycle(), petersen(). The actual body of these procedures
is normally not shown since the package initialization routine issues the
command
interface(verboseproc=0);. Once created, the graph can be modified by
using
various commands. To create a copy of a graph use the command
duplicate().
Further modifications of the original graph will not affect any such
copy.
- To generate a random graph use random(). Either the number of vertices
and
the probability of an edge occurring, or the number of vertices and
edges may
be specified.
- Vertices and edges can be added to the graph using the commands
addvertex(),
and addedge() . Vertex names are numeric by default but may be any
valid
maple string. They have a default weight of 0. Edges names are taken
from
the sequence e.(1..m) by default, but may be any string beginning with
the
letter `e`. Edges may be specified as sets or lists of vertices. A
list
indicates a directed edge while a set indicates an undirected edge.
Edges
have a default weight of 1. Weights may be general Maple expressions.
- All deletions are handled by the delete() command.
- To determine properties of a given graph G use commands such as
edges(G), or
vertices(G).
- The functions available are:
addedge
arrivals
bicomponents
connectivity
contract
daughter
degreeseq
djspantree
draw
fundcyc
incidence
mincut
mindegree
petersen
random
spantree
vweight
acycpoly
tuttepoly
charpoly
addvertex
adjacency
allpairs
ancestor
complement
complete
components
connect
countcuts
counttrees
cycle
cyclebase
delete
departures
diameter
dinic
duplicate
getlabel
incident
edges
girth
indegree
ends
graphical
induce
eweight
gsimp
maxdegree
neighbors
new
outdegree
path
rank
tail
shortpathtree
vdegree
show
vertices
shrink
void
chrompoly
flowpoly
rankpoly
spanpoly
flow
head
span
gunion
- The routines above include implementations of a number of basic
routines
including network flows, connectivity, disjoint spanning trees, allpairs
shortest path, single-source shortest path, minimum weight spanning
tree,
Tutte polynomials and special evaluations of them (rank, span,
acyclicity,
chromatic and flow polynomials), characteristic polynomials.
EXAMPLES:
> with(networks):
> new(G):
> addvertex({1,2,3,4},G);
1, 2, 3, 4
> connect({1,2},{3,4},G);
e1, e2, e3, e4
> edges(G);
{e1, e2, e3, e4}
> connectivity(G);
2
> countcuts(G);
6
> draw(G):
SEE ALSO: with, networks[<function>] (where <function> is any of the
functions
in the networks package
-----------------------------------------------------------------Here is the output of the command ?networks(spantree):
FUNCTION: networks[spantree] - finds a minimum weight spanning tree
CALLING SEQUENCE:
spantree(G);
spantree(G,s);
spantree(G,s,w);
PARAMETERS:
G - A graph or network
s - The starting or root vertex for the tree
w - a name for returning the sum of the edge weights in the tree
SYNOPSIS:
- This routine constructs a spanning tree for the graph G. The result is
returned as a new graph derived from G and consisting of that spanning
tree.
- It fails if G is not connected.
- The chosen tree has edges which minimize the total edge weight of the
tree.
- The routine is normally loaded via the command with(networks) but may
also be
referenced using the full name networks[spantree](...).
EXAMPLES:
> with(networks):
> G:=petersen():
> ends(G);
{{5, 9}, {1, 6}, {2, 8}, {3, 10}, {4, 7}, {1, 2}, {2, 3}, {3, 4}, {4,
5},
{1, 5}, {6, 7}, {7, 8}, {8, 9}, {9, 10}, {6, 10}}
> spantree(G,1,w):
> ends(");
{{5, 9}, {1, 6}, {2, 8}, {3, 10}, {4, 7}, {1, 2}, {4, 5}, {8, 9}, {6,
10}}
> w;
9
SEE ALSO: with, networks, shortpathtree, induce