�
advertisement

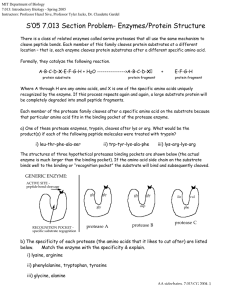
MIT Biology Department 7.012: Introductory Biology - Fall 2004 Instructors: Professor Eric Lander, Professor Robert A. Weinberg, Dr. Claudette Gardel� 7.012 Section Problem- Enzymes/Protein Structure There is a class of related enzymes called serine proteases that all use the same mechanism to cleave peptide bonds. Each member of this family cleaves protein substrates at a different location – that is, each enzyme cleaves protein substrates after a different specific amino acid. Formally, they catalyze the following reaction. A-B-C-D-X-E-F-G-H + H2O --------------->A-B-C-D-X protein substrate protein fragment + E-F-G-H protein fragment Where A through H are any amino acids, and X is one of the specific amino acids uniquely recognized by the enzyme. If this process repeats again and again, a large substrate protein will be completely degraded into small peptide fragments. Each member of the protease family cleaves after a specific amino acid on the substrate because that particular amino acid fits in the binding pocket of the protease enzyme. a) One of these protease enzymes, trypsin, cleaves after lys or arg. What would be the product(s) if each of the following peptide molecules were treated with trypsin? i) leu-thr-phe-ala-ser ii) trp-tyr-lys-ala-phe iii) lys-arg-lys-arg The structures of three hypothetical proteases binding pockets are shown below (the actual enzyme is much larger than the binding pocket). If the amino acid side chain on the substrate binds well to the binding or “recognition pocket” the substrate will bind and subsequently cleaved. GENERIC ENZYME: ACTIVE SITE -� peptide bond cleavage� gly gly gly RECOGNITION POCKET -� specific substrate regognition� protease A gly gly asp protease B ile val phe protease C b) The specificity of each protease (the amino acids that it likes to cut after) are listed below. Match the enzyme with the specificity & explain. i) lysine, arginine ii) phenylalanine, tryptophan, tyrosine iii) glycine, alanine AA sidechains, 7.013 CG 2004, 1 c) How might you design a similar enzyme to cleave after aspartic acid? d) Speculate on the effect of changing the aspartic acid in protease B to a glutamic acid. e) There are three amino acids required for the active site to function and three amino acids involved in substrate recognition – why then do these enzymes typically contain more than 200 amino acids? f) Suppose you make a solution of protease A. A small sample taken when the solution was made (time = 0) is capable of cleaving 100 mmol of protein substrate per minute. This rate of substrate cleavage is called the “activity” of the enzyme. At regular intervals over the next few hours as the solution of protease A stands at room temperature, you remove small samples and measure the activity (the rate at which a substrate protein is cleaved.). You find that the activity of the enzyme drops rapidly as time passes. However, interestingly, if when you made your solution of protease A, you added a large amount of casein (a protein found in milk that has no enzymatic activity), the protease loses its activity much more slowly. These data are sketched below: activity 100 protease A + casein protease A alone 0 0 time Explain these observations. AA sidechains, 7.013 CG 2004, 2 g) If you monitor the reaction of any of these proteases as they degrade a protein, you observe that the protease does not cut all the recognition sites in the substrate at once – the recognition sites on the surface of the substrate protein are the first to be cut. Explain. h) How might your answer to parts (f) and (g) be combined to design a long - lasting protease A molecule? i) Normally, protease A is synthesized as an inactive precursor, “zymogen A”. Zymogen A can be cleaved by protease A to form active protease A. catalyzed by protease A zymogen A + H2O -----------------------------> protease A + small peptide (inactive) (active) Draw a graph of the activity (see part (f)) as a function of time for a solution of zymogen A after the addition of a small amount of protease A. AA sidechains, 7.013 CG 2004, 3 at pH 7.0 STRUCTURES OF AMINO ACIDS GENERIC AMINO ACID: O O Individual amino acids are linked through these groups to form the backbone of the protein. O O H NH3 + C CH2CH2CH2 N C CH2 SH H H O H N C + H� C CH2� NH3 +� O H C� N C H� O C S CH3 H NH3� + H H� C C CH3 C CH3 H C CH2 C NH3 OH + H CH3 O O H� C CH2 H H N CH2 H + H H H C CH2 SERINE� (ser)� O O C C CH2 OH H OH NH3� + H NH3 + H NH3+ O� C� C H H TRYPTOPHAN (trp) O O H H CH2 PROLINE (pro) H N C CH2CH2CH2CH2 LYSINE (lys)� O H O C� NH3� + C NH3 + THREONINE (thr) H C CH2 H C CH2 O C� GLYCINE (gly) LEUCINE (leu) H C H NH3� +� O C PHENYLALANINE (phe) O H NH2 NH3 +� O C� NH3 +� METHIONINE� (met)� O O C� H H ISOLEUCINE (ile) O� C O O NH3 CH3 + HISTIDINE (his)� C CH2CH2 O C H O C GLUTAMINE (glN) H� C C CH2CH3 H C CH2CH2 O ASPARTIC ACID� (asp)� O NH3 + GLUTAMIC ACID (glu)� CYSTEINE (cys) H C NH3 + O C O O� C CH2 C NH2 O O C CH2CH2 H ASPARAGINE (asN) O� NH3 + O NH3 + NH2 + C NH3� + O C C O H� C CH2 C NH2 O� O C H O Peptide bonds O O ARGININE (arg) O C� O O NH3 + ALANINE (ala) H Side chain, unique to each differnt� amino acid NH3 + C H� C CH3 O H� C R O C O H R1 H O H R 3 C C N C C C N N C H O H R2 H� Protein Synthesis C H TYROSINE (tyr) H CH3� C C NH3 H + CH3 VALINE� (val)� S S GLY ALA VAL LEU ILE SER THR ASP ASN GLU GLN LYS ARG HIS TRP PHE TYR PRO CYS MET G A V L I S T D N E Q K Figure by MIT OCW. R H W F Y P C M AA sidechains, 7.013 CG 2004, 4